The Fluorescent Protein Color Palette
A broad range of fluorescent protein genetic variants have been developed over the past several years that feature fluorescence emission spectral profiles spanning almost the entire visible light spectrum. Extensive mutagenesis efforts in the original jellyfish protein have resulted in new fluorescent probes that range in color from blue to yellow and are some of the most widely used in vivo reporter molecules in biological research. Longer wavelength fluorescent proteins, emitting in the orange and red spectral regions, have been developed from the marine anemone Discosoma striata and reef corals belonging to the class Anthozoa. Still other species have been mined to produce similar proteins having cyan, green, yellow, orange, red, and far-red fluorescence emission. Developmental research efforts are ongoing to improve the brightness and stability of fluorescent proteins, thus improving their overall usefulness.
Essential to the understanding of spectral diversity in the wide range of fluorescent proteins discovered thus far are structural investigations of the stereochemical nature of the fluorophore and the effects of its surrounding environment on fluorescent properties. Aside from the jellyfish proteins, there appears to be a high degree of variation in the fluorophores of red-shifted fluorescent proteins. Even through the DsRed fluorophore configuration, termed planar cis, appears to be the predominant structure in most proteins that emit in the orange and red regions, there are at least two additional motifs, planar trans and non-planar trans, which have been elucidated through x-ray diffraction studies. A planar trans motif is found in the red fluorescent protein eqFP611, isolated from Entacmaea quadricolor, which displays one of the largest Stokes shifts and red-shifted emission wavelength profiles of any naturally occurring Anthozoan fluorescent protein. In contrast, the non-planar trans conformation is characteristic of the non-fluorescent chromoprotein Rtms5 from Montipora efflorescens. Several of the new mFruit proteins, described below, feature unusual chromophore architecture that deviates significantly from a strictly planar geometry.
A number of guiding motifs have emerged in recent years regarding the fundamental origins and manipulation of fluorescent protein emission color. The most important consideration appears to be the physical extent of p-orbital conjugation contained within the chromophore, which largely determines the general spectral class (such as cyan, green, yellow, or red). In addition, local environmental variables, such as the position of charged amino acid residues, hydrogen bonding network, and hydrophobic interactions within the protein matrix, are capable of producing either blue or red spectral shifts in the absorption and emission maxima by as much as 20 nm. As further studies into the complex characteristics of fluorescent protein chromophores yield clues about the structure-function relationship with the polypeptide backbone, the task of genetically engineering more finely-tuned color variants and broadening the spectral range of useful proteins will undoubtedly become easier.
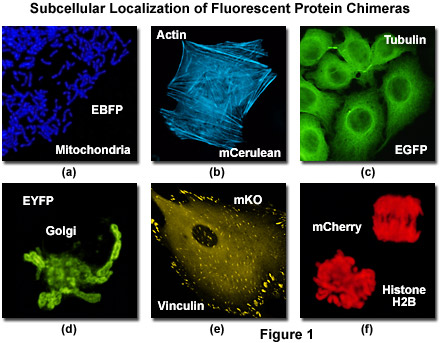
Illustrated in Figure 1 is the localization of a variety of fluorescent protein fusion tags to specific subcellular compartments. The chimera of enhanced blue fluorescent protein (EBFP) and the targeting sequence from subunit VIII of human cytochrome c oxidase histone localizes to the mitochondria (Figure 1(a)) in cultured human cells, while a similar fusion of mCerulean (a cyan fluorescent protein) and human beta-actin highlights the filamentous actin cytoskeletal network (Figure 1(b)) in African green monkey fibroblasts. The classical and most commonly employed fluorescent label, enhanced green fluorescent protein (EGFP) forms a similar cytoskeletal network when paired with alpha-tubulin (Figure 1(c)), and the yellow variant (EYFP) can be fused to the N-terminal 81 amino acids of human beta-1,4-galactosyltransferase to localize a yellow-green fluorescent marker to the Golgi apparatus (Figure 1(d)). Focal adhesions become fluorescent with a chimera of mKusabira Orange fluorescent protein (mKO; Figure 1(e)) and vinculin in fox lung fibroblasts, and the monomeric red fluorescent protein mCherry highlights the nucleus and condensed chromosomes when fused to histone H2B (Figure 1(f)) in human cervical carcinoma cells.
Blue Fluorescent Proteins
Fluorescent proteins emitting in the blue region of the visible light spectrum (approximately 440 to 470 nanometers) were first obtained from site-directed mutagenesis efforts targeted at the tyrosine amino acid residue at position 66 in the GFP chromophore (see Figure 2). Conversion of this residue to histidine (Y66H) produces a blue fluorescent protein (BFP) that exhibits a broad absorption band in the ultraviolet centered close to 380 nanometers and an emission maximum at 448 nanometers. The original protein exhibited only about 15 to 20 percent of the parent GFP brightness value due to a low quantum yield and required additional secondary mutations to increase its folding efficiency and expression levels. Subsequent investigations and several additional mutations led to an enhanced BFP version that is still only 25 percent as bright as the enhanced green variant and displays limited photostability compared to many other fluorescent proteins.
The primary motivation for developing blue fluorescent proteins in the mid-to-late 1990s was the keen interest in creating matched pairs for fluorescence resonance energy transfer (FRET) experiments and multicolor labeling. Because the spectral characteristics (fluorescence emission profile) of BFP are readily distinguishable from EGFP, this protein combination was one of the first utilized for multicolor imaging. Blue fluorescent protein also has the distinction of being incorporated into the first genetically encoded biosensor along with an enhanced GFP variant to demonstrate FRET through linkage of the two fluorescent proteins via an intervening protease-sensitive spacer. The broad emission peak of BFP overlaps to a significant extent with the excitation spectrum of red-shifted GFP variants to yield a Förster distance of 4.1, a reasonable value for measuring FRET. Blue fluorescent protein has also been coupled with several GFP derivatives into biosensors designed to monitor transcription factor dimerization, calcium, and apoptosis.
Aside from the limited brightness levels and rapid photobleaching, blue fluorescent proteins also suffer from the fact that they must be excited with ultraviolet light, which is phototoxic to mammalian cells, even in limited doses. Furthermore, working in this spectral region is often hampered by autofluorescence and high absorption levels by cells and tissues, as well as light scattering. Microscopes operating in the ultraviolet also require specialized light sources, optics, and filter combinations that further complicate imaging. For all of the reasons listed above, the quest for more efficient blue fluorescent proteins has only been pursued by a few research groups. Investigations of mutagenesis using non-natural amino acids positioned in and around the chromophore have led to several blue-shifted "artificial" fluorescent protein variants that may find utility in several biological and photophysical applications.
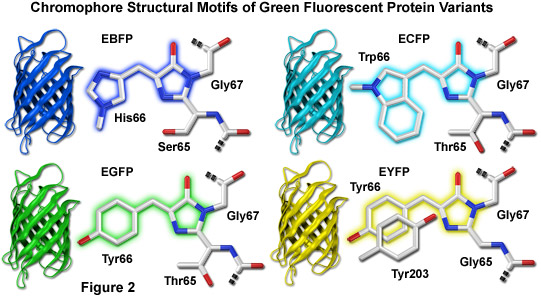
Illustrated in Figure 2 are the chromophore structures for color variants of green fluorescent protein. In all cases, the first step in maturation of the chromophore is a series of torsional adjustments that relocate the carboxyl carbon of the amino acid at position 65 so that it is in close proximity to the amino nitrogen of the glycine residue at position 67 (Gly67) in the polypeptide backbone. Nucleophilic attack by this carbon atom on the amide nitrogen of glycine, followed by dehydration, results in formation of an imidazolin-5-one heterocyclic ring system. Fluorescence occurs when oxidation of the aromatic amino acid (position 66) carbon bond by molecular oxygen extends electron conjugation of the imidazoline ring system to include the aromatic substituent. The cyan, green, and yellow fluorescent protein variant chromophores are discussed in greater detail in the following sections.
Cyan Fluorescent Proteins
The first fluorescent protein emitting in the bluish-green cyan spectral region (CFP; ranging from approximately 470 to 500 nanometers) was discovered simultaneously with BFP during mutagenesis studies that converted the tyrosine residue in the GFP chromophore to tryptophan (Y66W). This single mutation yielded a chromophore that displays an absorption maximum at 436 nanometers with a very broad fluorescence emission spectral profile centered at 485 nanometers. Subsequent refinements, including the F64L maturation improvement and S65T (discussed in the Green Fluorescent Protein section), resulted in the production of an enhanced version (ECFP; see Figure 2) with greater brightness and photostability. Even these modifications failed, however, to increase the brightness level of ECFP beyond 40 percent of that shown by EGFP. Other than providing an additional hue for multicolor imaging, initially the most promising aspect of ECFP was the potential for utility as a biosensor FRET partner with yellow fluorescent proteins in widefield and confocal microscopy using blue-violet filter sets or the 457-nanometer spectral line of an argon-ion laser. Greater photostability compared to EBFP also positions ECFP as a more useful protein for time-lapse imaging in localization and dynamics studies.
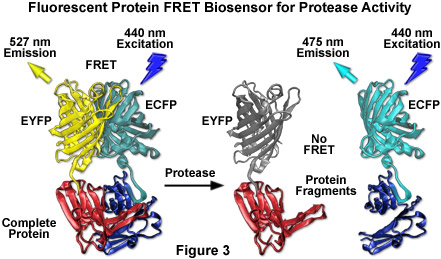
By far the most successful applications using ECFP have taken advantage of the ability of this variant to be coupled with yellow fluorescent protein (YFP) and derivatives to generate FRET biosensors capable of monitoring a wide spectrum of intracellular processes (see Figure 3). Although a host of FRET investigations have been performed using the ECFP-EYFP pair, the experimental results are often problematic due to limitations in the fluorophore properties that restrict measurements to a small dynamic range. A majority of these biosensors exhibit a typical overall ratio change of 10 to 30 percent during FRET analysis, with a few exceptions. Such a low level of contrast presents difficulties with modern digital microscopy due to a noise level that can approach 10 percent of the signal at low imaging intensities. These biosensors are further complicated by the potential to form dimers when constrained with tight two-dimensional spatial restrictions such as those that exist in membranes. To overcome the artifacts associated with dimerization, hydrophobic residues at the dimer interface were replaced with positively charged amino acids in both ECFP and EYFP to produce true monomeric variants (mCFP and mYFP), which achieve a far higher level of efficiency in FRET experiments. The critical mutation is the substitution of lysine for alanine at position 206 (A206K), which can be applied to virtually any Aequorea victoria variant in order to generate a true monomer capable of superior performance in applications such as localization, FRET, and protein dynamics.
Despite the host of improvements in CFP and the large database of experiments performed with this variant, continued investigation has produced additional useful fluorescent proteins in the cyan spectral class. Among the improved cyan fluorescent proteins that have recently been introduced, CyPet and an enhanced cyan variant termed Cerulean show the most promise as fusion tags, FRET biosensors, and for multicolor imaging (see Figures 1, 3, and 11). The Cerulean fluorescent probe (named for the sky-blue color) was rationally engineered by site-directed mutagenesis of enhanced cyan fluorescent protein to yield a higher extinction coefficient, improved quantum yield, and a fluorescence lifetime decay having a single exponential component (useful in lifetime decay measurements of FRET). Cerulean is at least 2-fold brighter than enhanced cyan fluorescent protein and has been demonstrated to significantly increase contrast as well as the signal-to-noise ratio when coupled with yellow-emitting fluorescent proteins, such as Venus (discussed below), in FRET investigations. In addition to site-directed mutations designed to improve folding and brightness, random mutagenesis was conducted on pre-Cerulean variants to further increase the molar extinction coefficient, and the optimized protein has been "monomerized" to enhance its utility in fusions. The abundance of advantageous features afforded by Cerulean render this protein the most useful all-purpose cyan derivative.
The cyan fluorescent protein variant referred to as CyPet (the acronym for: Cyan fluorescent Protein for energy transfer) was derived through a quantitative and unique strategy utilizing fluorescence-activated cell sorting to enhance the cyan and yellow pairing for FRET. Note that in designing probes for FRET investigations, optimization of the donor quantum yield and the acceptor extinction coefficient, as well as the overlap integral between these variables, are among the most critical parameters for achieving improved efficiency. Libraries of CFP and YFP variants were screened for FRET efficiency and the best clones were subjected to several evolutionary cycles consisting of random mutagenesis and synthetic DNA shuffling. A total of seven mutations resulted in the production of the CyPet protein, which features absorption and emission maxima positioned at 435 and 477 nanometers, respectively. CyPet is about half as bright as EGFP and two-thirds as bright as Cerulean, but expresses relatively poorly at 37 degrees Celsius. However, when paired with its FACS-optimized partner, YPet, in FRET biosensors, this cyan variant exhibits a dynamic range that is more than six times higher than CFP-YFP, dramatically improving contrast and potentially leading to far more sensitive detection of subtle intracellular processes. CyPet has a more blue-shifted and narrower fluorescence emission peak than mCFP, which greatly increases its utility in multicolor imaging, and is the most photostable cyan fluorescent protein of the weakly dimeric and monomeric versions currently available.
Several potentially useful cyan proteins have been isolated in Anthozoan species. Derived from the reef coral Anemonia majano, the AmCyan1 fluorescent protein, which is now commercially available (Clontech), has been optimized with human codons for enhanced expression in mammalian cell systems. Originally named amFP486 (am, Anemonia majano; FP, fluorescent protein; 486 emission maximum) in accordance with a nomenclature scheme devised to simplify the discussion of myriad Anthozoan proteins, this variant exhibits a similar brightness level, but a significantly better resistance to photobleaching than CFP. The absorption maximum of AmCyan1 occurs at 458 nanometers while the fluorescence emission peak resides at 489 nanometers. Note that both peaks are shifted to longer wavelengths by 19 and 13 nanometers, respectively, compared to ECFP. On the downside, similar to most of the other reef coral proteins, the probe has a tendency to form tetramers, which will significantly complicate attempts to employ this protein as a fusion tag or a FRET biosensor.
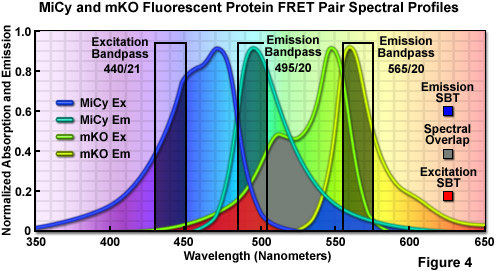
First isolated by Miyawaki and associates from an Acropara stony coral species, the cyan-emitting Midori-Ishi Cyan (abbreviated MiCy) probe was originally designed as the donor in a new FRET combination with the monomeric Kusabira Orange (mKO) fluorescent protein to generate a biosensor probe with high spectral overlap (Förster distance of 5.3; see Figure 4; mKO is discussed in the section on Orange Fluorescent Proteins). This protein features the longest absorption and emission wavelength profiles (472 and 495 nanometers, respectively) reported for any probe in the cyan spectral class. The high molar extinction coefficient and quantum yield exhibited by MiCy render the protein of equal brightness to Cerulean, although the spectra are far more sensitive to pH. Also similar to Cerulean, MiCy features a single exponential lifetime decay component with a time constant of 3.4 nanoseconds, which should be useful for measurements of FRET in combination with fluorescence lifetime imaging microscopy (FLIM). An unusual feature of MiCy is that it forms a homodimeric complex similar to the GFP variant isolated from the bioluminescent sea pansy, Renilla reniformis, rather than the obligate tetramer observed in most coral reef species. Although the dimerization motif may be a problem in some fusion proteins, it should be far easier than a tetramer to mutate MiCy into a true monomer.
Recently, a new monomeric cyan fluorescent protein having superior brightness, insensitivity to acidic conditions, and photostability has been introduced for live-cell imaging applications of fusion partners, and as a FRET donor for yellow and orange acceptor fluorescent proteins in biosensors. Termed mTFP1 (monomeric teal fluorescent protein 1), the variant was produced from a synthetic gene library built around the tetrameric cyan protein, cFP484, originating from a Clavularia soft coral. Displaying red-shifted spectral profiles (excitation and emission maxima at 462 and 492 nanometers, respectively) when compared to other cyan members of this spectral class, mTFP1 has a total of 31 amino acid substitutions relative to the wild-type tetramer. The red-shifted spectra were considered when classifying the new color as teal rather than cyan. Unlike other members of the cyan fluorescent protein group, which generally feature the aromatic amino acid tryptophan at position 66 in the chromophore, mTFP1 contains the classical tyrosine residue at this location, characteristic of many GFP derivatives. Substituting tyrosine for tryptophan reduces the broad fluorescence emission spectral width from approximately 60 nanometers to a narrower and more manageable 30 nanometers, a factor that is useful for reducing bleed-through in multi-color experiments. The high quantum yield (0.85) of mTFP1 provides an excellent alternative to the cyan derivatives, mCFP and mCerulean, as a FRET donor having a Förster distance exceeding 5.0 when combined with either yellow or orange fluorescent proteins.
Illustrated in Figure 4 are the absorption and emission spectral profiles of MiCy and mKO along with recommended bandpass filters for excitation and gathering of fluorescence emission for FRET analysis using the two probes. The overlap region between the emission spectrum of MiCy and the absorption spectrum of mKO is presented with a gray fill, while the excitation and emission spectral bleed-through regions are shown in red and blue fills, respectively. Applying an excitation bandpass filter centered at 440 nanometers with a 21-nanometer bandwidth reduces the level of excitation of mKO, and the 495/20-nanometer emission filter enables analysis of the donor emission (MiCy) fluorescence without contamination of signal from the acceptor (mKO). The 565/20-nanometer acceptor bandpass filter encompasses the maximum emission region of mKO with approximately 7-10 percent bleed-through of the MiCy signal. A dichromatic mirror having a cut-on wavelength of 485 nanometers (not illustrated) should be utilized with this filter combination.
Green Fluorescent Proteins
The original green fluorescent protein isolated from Aequorea victoria (termed wild-type) has been the principal subject of numerous investigations, but is not useful in a majority of the practical applications involving fluorescent proteins due to the bimodal absorption band (395 and 475 nanometers peaks), which is hampered by relatively low extinction coefficients and a principal absorption maximum residing in the ultraviolet part of the spectrum. Shortly after GFP was demonstrated to be a useful marker for gene expression, a point mutation altering the serine residue at position 65 in the chromophore into threonine (S65T) produced a new version of the protein having a well defined absorption profile with a single peak at 484 nanometers. This mutation is featured in the most popular variant of green fluorescent protein, termed enhanced green fluorescent protein (EGFP), which can be imaged using commonly available filter sets designed for fluorescein (FITC) and is among the brightest and most photostable of the Aequorea victoria fluorescent proteins. These features have rendered enhanced green fluorescent protein one of the most popular probes and the best choice for most single-label live-cell fluorescence imaging experiments. As previously discussed, EGFP was coupled with blue fluorescent proteins as a FRET acceptor in early biosensor investigations, but has subsequently been largely replaced by yellow, orange, and red variants. The only drawbacks to the use of EGFP as a fusion tag are a slight sensitivity to pH and a weak tendency to dimerize.
In addition to EGFP, several other green-emitting variants (in the range of approximately 500 to 525 nanometers) are currently being utilized in live-cell imaging experiments. Among the best of these in terms of photostability and brightness may be the Emerald variant, but lack of a commercial source (until recently) has limited its use. Emerald contains the S65T mutation featured in EGFP, but also has four additional point mutations that improve folding, expression at 37 degrees Celsius, and brightness. Although Emerald is far more efficient than EGFP in folding and developing fluorescence in mammalian cells, it has a fast photobleaching component that might affect quantitative imaging in some environments. One of the most interesting derivatives of wtGFP is Sapphire, which contains a critical mutation of isoleucine for threonine at position 203 (T203I), abolishing the absorption peak at 475 nanometers. The result is a protein having an absorption maximum at 399 nanometers with emission in the green spectral region (511 nanometers) to yield a surprisingly large Stokes shift of over 100 nanometers. Several Sapphire variants have been developed to improve folding, including a probe known as T-Sapphire (for Turbo), and derivatives featuring circular permutations are available that might be useful for altering the orientation geometry with fusion partners. Due to the significant separation between absorption and emission peaks, the greatest potential for the Sapphire proteins is their pairing with orange and red derivatives (see Figure 8) as FRET donors.
A wide variety of additional fluorescent proteins emitting in the green spectral region have been isolated from other sources, including different Aequorea species, copepods, and coral reefs. One of the most promising of these probes, derived by random mutagenesis of a colorless protein isolated from Aequorea coerulescens, is known as aceGFP. The conversion of glutamic acid into glycine at position 222 (E222G) transformed the wild-type aceGFP protein into a highly fluorescent species with a relatively symmetrical pair of spectral profiles having an absorption maximum at 480 nanometers and an emission peak at 505 nanometers. The high molar extinction coefficient and quantum yield of aceGFP combine to produce a brightness level similar to that displayed by EGFP. Demonstrated to exist in aqueous solution as a monomer by electrophoresis and gel filtration, this protein is commercially available from several sources (Clontech and Evrogen, as AcGFP1 and AceGFP, respectively) with human-optimized codon replacements. Proper localization of fusion products targeted at specific subcellular components and organelles (such as filamentous actin, the Golgi, nucleus, and mitochondria; see Figure 1) indicates that aceGFP is quite useful as a marker and could have potential for partnership with red-emitting proteins in a novel FRET combination. However, the photostability characteristics of aceGFP remain unknown, and there are no clear advantages to the use of this protein over the more common EGFP and Emerald variants.
Fluorescent Protein Properties
| Protein (Acronym) |
Excitation Maximum (nm) |
Emission Maximum (nm) |
Molar Extinction Coefficient |
Quantum Yield |
in vivo Structure |
Relative Brightness (% of EGFP) |
| GFP (wt) | 395/475 | 509 | 21,000 | 0.77 | Monomer* | 48 |
| Blue Fluorescent Proteins | ||||||
| EBFP | 383 | 445 | 29,000 | 0.31 | Monomer* | 27 |
| Sapphire | 399 | 511 | 29,000 | 0.64 | Monomer* | 55 |
| T-Sapphire | 399 | 511 | 44,000 | 0.60 | Monomer* | 79 |
| Cyan Fluorescent Proteins | ||||||
| ECFP | 439 | 476 | 32,500 | 0.40 | Monomer* | 39 |
| mCFP | 433 | 475 | 32,500 | 0.40 | Monomer | 39 |
| Cerulean | 433 | 475 | 43,000 | 0.62 | Monomer* | 79 |
| CyPet | 435 | 477 | 35,000 | 0.51 | Monomer* | 53 |
| AmCyan1 | 458 | 489 | 44,000 | 0.24 | Tetramer | 31 |
| Midori-Ishi Cyan | 472 | 495 | 27,300 | 0.90 | Dimer | 73 |
| mTFP1 (Teal) | 462 | 492 | 64,000 | 0.85 | Monomer | 162 |
| Green Fluorescent Proteins | ||||||
| EGFP | 484 | 507 | 56,000 | 0.60 | Monomer* | 100 |
| AcGFP | 480 | 505 | 50,000 | 0.55 | Monomer* | 82 |
| TurboGFP | 482 | 502 | 70,000 | 0.53 | Monomer* | 110 |
| Emerald | 487 | 509 | 57,500 | 0.68 | Monomer* | 116 |
| Azami Green | 492 | 505 | 55,000 | 0.74 | Monomer | 121 |
| ZsGreen | 493 | 505 | 43,000 | 0.91 | Tetramer | 117 |
| Yellow Fluorescent Proteins | ||||||
| EYFP | 514 | 527 | 83,400 | 0.61 | Monomer* | 151 |
| Topaz | 514 | 527 | 94,500 | 0.60 | Monomer* | 169 |
| Venus | 515 | 528 | 92,200 | 0.57 | Monomer* | 156 |
| mCitrine | 516 | 529 | 77,000 | 0.76 | Monomer | 174 |
| YPet | 517 | 530 | 104,000 | 0.77 | Monomer* | 238 |
| PhiYFP | 525 | 537 | 124,000 | 0.39 | Monomer* | 144 |
| ZsYellow1 | 529 | 539 | 20,200 | 0.42 | Tetramer | 25 |
| mBanana | 540 | 553 | 6,000 | 0.70 | Monomer | 13 |
| Orange and Red Fluorescent Proteins | ||||||
| Kusabira Orange | 548 | 559 | 51,600 | 0.60 | Monomer | 92 |
| mOrange | 548 | 562 | 71,000 | 0.69 | Monomer | 146 |
| dTomato | 554 | 581 | 69,000 | 0.69 | Dimer | 142 |
| tdTomato (Tandem) | 554 | 581 | 138,000 | 0.69 | Monomer | 283 |
| DsRed | 558 | 583 | 75,000 | 0.79 | Tetramer | 176 |
| DsRed2 | 563 | 582 | 43,800 | 0.55 | Tetramer | 72 |
| DsRed-Express (T1) | 555 | 584 | 38,000 | 0.51 | Tetramer | 58 |
| DsRed-Monomer | 556 | 586 | 35,000 | 0.10 | Monomer | 10 |
| mTangerine | 568 | 585 | 38,000 | 0.30 | Monomer | 34 |
| mStrawberry | 574 | 596 | 90,000 | 0.29 | Monomer | 78 |
| AsRed2 | 576 | 592 | 56,200 | 0.05 | Tetramer | 8 |
| mRFP1 | 584 | 607 | 50,000 | 0.25 | Monomer | 37 |
| JRed | 584 | 610 | 44,000 | 0.20 | Dimer | 26 |
| mCherry | 587 | 610 | 72,000 | 0.22 | Monomer | 47 |
| HcRed1 | 588 | 618 | 20,000 | 0.015 | Dimer | 1 |
| mRaspberry | 598 | 625 | 86,000 | 0.15 | Monomer | 38 |
| HcRed-Tandem | 590 | 637 | 160,000 | 0.04 | Monomer | 19 |
| mPlum | 590 | 649 | 41,000 | 0.10 | Monomer | 12 |
| AQ143 | 595 | 655 | 90,000 | 0.04 | Tetramer | 11 |
| * Weak Dimer | ||||||
Table 1
Presented in Table 1 is a compilation of properties displayed by several of the most popular and useful fluorescent protein variants. Along with the common name and/or acronym for each fluorescent protein, the peak absorption and emission wavelengths (given in nanometers), molar extinction coefficient, quantum yield, relative brightness, and in vivo structural associations are listed. The computed brightness values were derived from the product of the molar extinction coefficient and quantum yield, and are divided by the value for EGFP to calculate the percentage documented in the table. This listing was created from scientific and commercial literature resources and is not intended to be comprehensive, but instead represents fluorescent protein derivatives that have received considerable attention in the literature and may prove valuable in research efforts. Furthermore, the absorption and fluorescence emission spectra listed in the table and illustrated in this review were recorded under controlled conditions and are normalized for comparison and display purposes only. In actual fluorescence microscopy investigations, spectral profiles and wavelength maxima may vary due to environmental effects, such as pH, ionic concentration, and solvent polarity, as well as fluctuations in localized probe concentration. Therefore, the listed extinction coefficients and quantum yields may differ from those actually observed under experimental conditions.
Several closely related GFP-like proteins have been isolated from an assortment of copepod aquatic crustacean species. The brightest of these probes, originally referred to as ppluGFP2, have been made commercially available (Evrogen) under the names CopGFP and TurboGFP (an enhanced variant). CopGFP is efficiently excited using an argon-ion laser or FITC blue excitation filter set (absorption maximum at 482 nanometers) and produces green fluorescence at 502 nanometers with a brightness value approximately 30 percent higher than EGFP and a much greater resistance to changes in pH. Reported to be a monomer in dilute solution, CopGFP matures significantly faster than EGFP and is ideal for applications as a fusion partner targeted at expression in subcellular regions of high acidity. Limitations of this probe include the inability to isolate stable cell lines and the formation of aggregates in long-term cultures. The improved version, TurboGFP, derived from site-directed and random mutagenesis, retains the fast maturation kinetics of the parent protein with a slight loss in brightness and substantially lower resistance to acidic environments. Despite the improved folding kinetics and excellent optical properties of these proteins, however, photostability data have not been reported and no compelling evidence exists to demonstrate a significant benefit over the application of the extensively studied original GFP derivatives.
Green fluorescent proteins have also been mined from reef corals and several of these are commercially available. A brightly fluorescent protein termed Azami Green, bearing only a surprisingly scant (less than 6 percent) sequence homology to EGFP, was isolated from the stony coral Galaxeidae and has been demonstrated to mature rapidly during expression in mammalian cell lines. Likewise, one of the original Anthozoa coral reef proteins from Zoanthus reported by Matz and coworkers has also been transformed into a commercial product (Clontech) under the tradename ZsGreen. The probes have absorption maxima at 492 and 496 nanometers and emission peaks at 505 and 506 nanometers, respectively, readily allowing visualization and imaging with standard lasers and filter combinations in confocal and widefield microscopy. However, similar to most of the other proteins isolated in corals, Azami Green and ZsGreen both exist as tetramers in the natural state, which significantly interferes with their use as fusion partners and as a FRET donor or acceptor in biosensors. To overcome the oligomerization problem, site-directed and random mutagenesis efforts were successful in creating a monomeric version of Azami Green, but this type of effort has not been reported for ZsGreen although the protein has been re-engineered with human codons to optimize expression (resulting in a variant termed ZsGreen1). Because reliable photostability data is lacking, it is unclear whether either of these proteins will outperform EGFP in long-term imaging experiments.
The sea pansy, an Anthozoa soft coral, is the source of several green fluorescent proteins that have been characterized in detail and are now commercially available. A protein isolated from Renilla reniformis that exhibits properties similar to EGFP is the best characterized of the probes in this class. Having absorption and emission maxima at 485 and 508 nanometers, respectively, in addition to a similar sensitivity to pH, the Renilla protein would be an excellent substitute for EGFP were it not for the fact that it is an obligate dimer. Aside from the oligomerization problem, Renilla GFPs may be useful in many applications and have been expressed in a wide variety of organisms, including bacteria, fungi, and mammalian cells. Versions with human codon sequences are available from the manufacturers, as are derivatives optimized for expression in other species. There is a general lack of reliable data concerning extinction coefficients, quantum yields, and photostability for the commercial Renilla proteins, so valid comparisons to EGFP in terms of brightness and photobleaching are not possible.
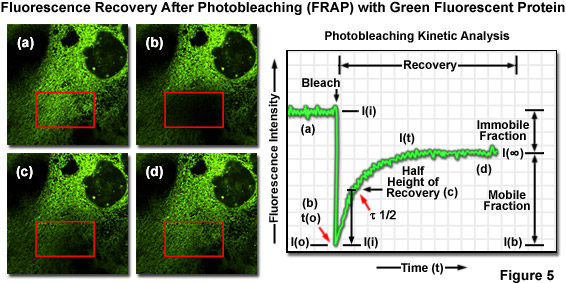
One of the most useful and popular live-cell imaging applications for green fluorescent protein and its derivatives is being a marker in fluorescence recovery after photobleaching (FRAP) experiments designed to investigate intracellular dynamics. Illustrated in Figure 5(a) is an epithelial cell labeled with EGFP fused to an endoplasmic reticulum targeting sequence that localizes the fluorescent probe to this organelle network. A region of interest (red box) is traced with an acousto-optic tunable filter (AOTF) in a laser scanning confocal microscope, and the area is then photobleached with high laser power for 5 seconds (Figure 5(b)), effectively quenching all of the fluorescence. Continued monitoring of the cell with an imaging laser (488-nanometer argon-ion) at low intensity for a much longer time period (several minutes to an hour) enables visualization of fluorescence recovery in the photobleached region (Figures 5(c) and 5(d)). A plot of fluorescence intensity versus time (Figure 5) enables quantitative analysis of the recovery kinetics and provides information about the diffusion coefficient and mobility for this fluorescent protein chimera.
Yellow Fluorescent Proteins
Yellow fluorescent proteins, as a spectral class, are among the most versatile genetically-encoded probes yet developed. Ranging in emission wavelength maxima from approximately 525 to 555 nanometers, those proteins residing in the shorter wavelength region actually appear green, rather than yellow, when viewed in a widefield fluorescence microscope. The first member in what has become a rather large family of probes was rationally engineered after the high resolution crystal structure of green fluorescent protein revealed that threonine residue 203 (Thr203) was positioned near the chromophore and potentially able to alter the spectral characteristics upon substitution. Mutations of this aliphatic amino acid to several aromatic moieties were introduced in order to induce pi-orbital stacking and attempt stabilization of the excited state dipole moment of the chromophore. The most successful mutant proved to be tyrosine (T203Y; termed mutant 10C, the original YFP), which resulted in almost a 20-nanometer shift to longer wavelengths for both the excitation and emission spectra. Several YFP variants were initially constructed in attempts to maximize brightness as well as to increase the speed of maturation and optimize expression at 37 degrees Celsius. One of these variants, named Topaz, has been of service in fusion tag localization, intracellular signaling, and FRET investigations.
In an effort to improve the performance of FRET biosensors, further sequence refinements led to the development of the enhanced yellow fluorescent protein (EYFP), which is one of the brightest and most widely utilized fluorescent proteins. EYFP was constructed from the original yellow variant by introduction of lysine to position 69 in place of glutamine (Q69K). The high brightness level and fluorescence emission spectrum wavelength profile of EYFP combine to make this probe an excellent candidate for multicolor imaging experiments in fluorescence microscopy, although maturation is slower, especially in organelles. Enhanced yellow fluorescent protein has also been widely employed as an acceptor for energy transfer experiments when paired with enhanced cyan fluorescent protein. However, all of the original yellow fluorescent protein variants present some problems in that they are very sensitive to acidic pH and lose approximately 50 percent of their fluorescence at pH 6.5. In addition, most of the Aequorea-derived proteins in this class have also been demonstrated to have significant sensitivity to chloride ions, undergo weak dimerization, exhibit relatively poor expression at 37 degrees Celsius, and to photobleach much more readily than many of the green fluorescent proteins. Several investigations have taken advantage of the environmental sensitivity of YFP to measure cytosolic pH, chloride ion concentrations, and FRET efficiency. The A206K mutation, substituting the charged amino acid lysine for alanine at the hydrophobic dimer interface, has been applied to YFP in order to generate a true monomer.
Continued genetic development of the YFP family led to the discovery that a single point mutation near the chromophore, the substitution of methionine for glutamine at position 69 (Q69M), dramatically increases the acid stability of the protein and reduces the chloride sensitivity. Named Citrine in recognition of the yellow color and acid resistance, this variant also expresses at a much higher level in mammalian cell culture (especially when targeted to acidic organelles) and demonstrates almost twice the photostability of many previous yellow fluorescent proteins. Citrine features absorption and fluorescence emission maxima at 516 and 529 nanometers, respectively, and is 75 percent brighter than EGFP, although much less photostable. FRET biosensors constructed with Citrine demonstrate better performance than sister analogs containing EYFP, and the probe is a better choice for multicolor experiments in general. Although Citrine exists as a weak dimer in solution, the protein can be converted into a monomer with the A206K mutation.
Introduction of yet another novel point mutation into Aequorea-derived YFP, the substitution of leucine for phenylalanine at position 46 (F46L), produced a variant that was named Venus in honor of the brightest star (or planet) in the night sky. This mutation, which was discovered during random mutagenesis experiments with circularly permuted GFP derivatives, dramatically accelerates oxidation of the chromophore, the rate-limiting step in fluorescent protein maturation. Additional mutations were also introduced in order to increase the tolerance of Venus to acidic environments and to reduce the sensitivity to chloride. The absorption and emission spectral peaks (515 and 528 nanometers, respectively) of Venus are shifted to longer wavelengths by a single nanometer compared to EYFP, but the brightness level is retained. Unfortunately, the photostability of Venus is only about 25 percent that of EYFP, a significant problem for long-term imaging experiments. Venus has been demonstrated to perform well in intracellular organelle fusion protein pH assays, for localization studies in yeast, in bimolecular fluorescence complementation analysis, and holds significant potential as an acceptor in FRET biosensors. Genetic conversion of the weakly dimeric Venus into a monomer using the A206K mutation will further expand the use of this probe in biological investigations.
During the same fluorescence-activated cell sorting investigation that led to the generation of the cyan fluorescent protein CyPet, the evolutionary optimized complementary FRET acceptor, termed YPet, was also obtained from Venus and YFP variants. Named after its proficiency in FRET (YFP for energy transfer), YPet is the brightest yellow fluorescent protein variant yet developed and demonstrates very good photostability. The resistance to acidic environments afforded by YPet is superior to Venus and other YFP derivatives, which will enhance the utility of this probe in biosensor combinations targeted at acidic organelles. However, although the optimized CyPet-YPet combination should be the preferred starting point in the development of new FRET biosensors, the utility of this pair has not yet been tested in widespread practice. Likewise, the suitability of CyPet and YPet in fusion tags for localization experiments, bimolecular complementation analysis, and other routine fluorescent protein assays has yet to be established. Both proteins exist in solution as weak dimers, but presumably can be converted to true monomers using the A206K mutation that has worked so well with other Aequorea variants.
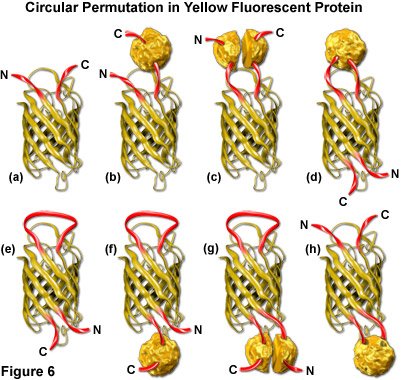
Both green and yellow fluorescent proteins have been genetically engineered to create circular permutations of the original sequences that enable fusions to amino acids far removed from the normal amino and carboxy termini (abbreviated cpGFP and cpYFP). The well-conserved beta-barrel structure of fluorescent proteins, coupled to the intricate post-translational chromophore maturation patterns exhibited by these probes, at first glance indicate that major insertions into the main polypeptide backbone would prevent fluorescence. However, several mutants have been developed (see Figure 6) that fuse the original carboxy (C) and amino (N) termini with a short spacer and create new locations within the barrel structure for the fusion of guest proteins. Although the nature of allowable fusions and the properties of the circular permutation variants are somewhat altered with respect to the standard fluorescent proteins, these new designs offer a unique opportunity to generate an original class of localization probes and physiological indicators.
The original EYFP molecule is represented as a cartoon drawing in Figure 6(a) to show the yellow barrel with the carboxy and amino termini represented as red ribbons. A tandem fusion product of EYFP with another protein (represented by a textured sphere) is diagrammed in Figure 6(b), and the insertion of EYFP into a split protein domain is illustrated in Figure 6(c). Relocation of the termini in circularly permuted EYFP to the barrel region with subsequent insertion of a protein at the original terminus location, and the same configuration minus the insertion protein are depicted in Figures 6(d) and 6(e), respectively. The last three cartoons in Figure 6 ((f), (g), and (h)) illustrate insertion of a fusion protein at the C-terminus (Figure 6(f)), insertion of split domains at both the C and N termini (Figure 6(g)), and insertion of a complete protein domain into the beta-barrel backbone of EYFP (Figure 6(h)). The first three cartoons in Figure 6 are standard fluorescent protein chimeras, while the last five are possibilities that could readily be constructed using circular permutation variants.
Although the potential for new discoveries of yellow and green fluorescent proteins in Hydrozoan species other than Aequorea victoria is significant, only one candidate has surfaced so far. Isolated from the Phialidium jellyfish, a protein termed phiYFP is reported to demonstrate very bright yellow fluorescence (absorption and emission at 525 and 537 nanometers, respectively) and to be useful for N-terminal fusion tags. An extraordinary feature of phiYFP is that the naturally occurring protein contains the same mutation at position 64 (leucine) that was introduced by Venus to increase the folding efficiency. The probe also naturally contains tyrosine at position 203, another site-directed modification of the native GFP that resulted in yellow fluorescence. This remarkable discovery of a natural similarity between the structure of phiYFP and genetically modified Aequorea proteins is a testament to the efficacy of protein engineering efforts directed at GFP to adjust the spectral properties. The phiYFP protein has been optimized by random mutagenesis to produce a monomeric version without compromising the spectral properties.
Site-directed and random mutagenesis of a monomeric Anthozoa fluorescent protein from Discosoma (mRFP1) have resulted in the creation of two monomeric coral reef derivatives with spectral properties falling in the range of Aequorea yellow fluorescent proteins. Named after similarly colored fruits, mHoneydew and mBanana both emit fluorescence in the yellow spectral region. The broad absorption band of mHoneydew (peaks at 487 and 504 nanometers) enables effective excitation with an argon-ion laser or standard FITC filter combination. However, the similarly broad emission spectrum (peaks at 537 and 562 nanometers) tails into the near-infrared, hampering the use of this protein in multicolor experiments. In addition, a low extinction coefficient and quantum yield render mHoneydew the dimmest member of the monomeric yellow fluorescent protein cadre. The mBanana variant is only twice as bright as mHoneydew, but features much narrower excitation and emission spectra (peaks at 540 and 553 nanometers, respectively). Because both proteins exhibit relatively poor photostability, and mBanana is highly pH-sensitive, neither will probably find great utility in imaging experiments. Perhaps the most promising aspect of these probes is that the mere existence of mHoneydew (a cyan-type Y67W mutant) demonstrates that the tryptophan-based chromophore of CFP can undergo a further maturation into a longer-wavelength emitting species.
ZsYellow (originally referred to as zFP538) is a yellow fluorescent protein that was discovered in the Anthozoan button polyp Zoanthus during a search in reef corals for naturally occurring GFP analogs emitting fluorescence in longer wavelength regions. One of the most unique features of the ZsYellow fluorescence emission spectrum is that the peak (538 nanometers) occurs almost midway between those of EGFP (508 nanometers) and DsRed (583 nanometers), presenting an opportunity to investigate proteins emitting fluorescence in the yellow portion of the visible light spectrum. The ZsYellow fluorescent protein chromophore features a novel three-ring system and peptide backbone cleavage due to the substitution of lysine for serine as the first amino acid residue in the chromophore tripeptide sequence. As a result of the unique chromophore motif, the degree of conjugation observed in ZsYellow is intermediate between that observed with EGFP and DsRed (one double bond more than EGFP, and one less than DsRed), which accounts for the positioning of emission wavelengths in the yellow region. ZsYellow exhibits a marked tendency to form tetramers when expressed in vivo, hampering the use of this protein as a fusion partner for localization investigations. Furthermore, the reduced brightness level of ZsYellow when compared to enhanced green fluorescent protein (25 percent of EGFP) also limits the utility of this reporter in fluorescence microscopy (the human codon-optimized version is commercially available as ZsYellow1). The unique emission spectral profile of ZsYellow, however, should encourage the search for genetic modifications that alleviate the tendency to form tetramers while simultaneously increasing the quantum yield and extinction coefficient, an effort that could ultimately yield a high-performance monomeric yellow fluorescent protein.
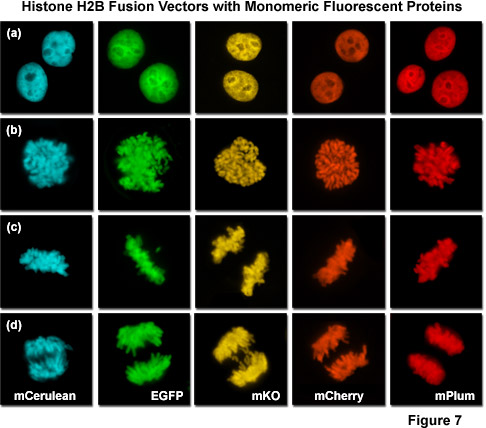
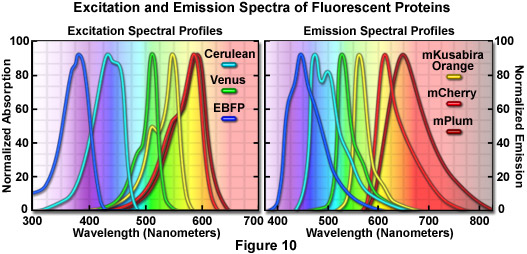
Presented in Figure 7 is a collage of histone H2B fusion proteins demonstrating the utility of the vast fluorescent protein color palette. Each chimera contains a selected monomeric fluorescent protein sequence (mCerulean, EGFP, mKO, mCherry, and mPlum) fused to the amino acid sequence for human histone H2B, separated with an intermediate linker unit containing six amino acids. The images in Figure 7 were collected from transiently transfected human cervical carcinoma (HeLa) cells after expression the fusion proteins for several days. Cells captured in various stages of mitosis are evident with all the chimeras. Interphase is illustrated for all of the proteins in panel (a) (horizontally), while prophase, metaphase, and anaphase are depicted in panels (b), (c), and (d), respectively. These fusion proteins are useful for mitosis investigations that require multicolor imaging with other fluorescent proteins. The wide spectrum of useful fluorescent protein mutants illustrated in Figure 7, which covers the entire visible spectral range (see Figure 10), provides a great deal of flexibility in the choice of imaging partners.
Orange Fluorescent Proteins
In contrast to the relatively large number of fluorescent proteins engineered in the cyan, green, and yellow spectral classes, only three probes have been developed so far in the orange portion of the spectrum (ranging from approximately 555 to 580 nanometers). Even so, all three of the existing orange fluorescent proteins, which were isolated from coral reef species, have the potential to be useful in a variety of imaging scenarios. Perhaps the most versatile of these is monomeric Kusabira Orange, a protein originally derived as a tetramer from the mushroom coral Fungia concinna (known in Japanese as Kusabira-Ishi). Kusabira Orange was engineered by site-specific mutagenesis from a cDNA clone of the coral by adding ten amino acids to the N-terminus. The resulting protein has an absorption maximum at 548 nanometers (ideal for excitation with a 543-nanometer laser) and emits bright orange fluorescence at 561 nanometers. A monomeric version of Kusabira Orange (abbreviated mKO) was created using a strategy similar to that employed for DsRed to create mRFP1 (discussed in the section on Red Fluorescent Proteins) by introducing over 20 mutations through site-directed and random mutagenesis. The monomer exhibits similar spectral properties to the tetramer and has a brightness value similar to EGFP, but is slightly more sensitive to acidic environments. The photostability of this probe, however, is the best of any fluorescent protein in all of the spectral classes, making mKO an excellent choice for long-term imaging experiments. Furthermore, the emission spectral profile is sufficiently well separated from cyan fluorescent proteins to increase the FRET efficiency in biosensors incorporating mKO, and the probe is useful in multicolor investigations (see Figure 4) with a combination of cyan, green, yellow, and red fluorescent proteins.

The mRFP1 derivative, mOrange, was derived after four rounds of directed evolution to yield a probe absorbing at 548 nanometers and emitting orange fluorescence at 562 nanometers. The mOrange variant is slightly brighter than mKusabira Orange, but has less than 10 percent the photostability, thus severely compromising its application for experiments that require repeated imaging. However, mOrange remains the brightest protein in the orange spectral class and is still an excellent choice where intensity is more critical than long-term photostability. In addition, when combined with the green-emitting T-Sapphire, mOrange is a suitable alternative to CFP-YFP proteins as a FRET pair to generate longer wavelength biosensors (Figure 8), and can be coupled with fluorescent proteins in other spectral regions for multicolor investigations. A novel orange fluorescent protein isolated from the Cerianthus tube anemone is commercially available (tradename cOFP; Stratagene) and has spectral properties that are similar to mOrange and mKusabira Orange, but like the other anemone proteins isolated to date, exists in solution as a tetramer. The brightness and photostability of cOFP have not been reported so this protein cannot be directly compared to other orange fluorescent proteins, and its utility will be further limited until it can be converted into a monomer.
The combination of Sapphire and mOrange fluorescent proteins as a FRET pair is presented in Figure 8. Illustrated are the absorption and emission spectral profiles of the two probes along with recommended bandpass filters for excitation and collecting fluorescence emission for FRET analysis. The overlap region between the emission spectrum of Sapphire and the absorption spectrum of mOrange is presented with a gray fill, while the excitation and emission spectral bleed-through regions are depicted in red and blue fills, respectively. Applying an excitation bandpass filter centered at 395 nanometers with a 30-nanometer bandwidth reduces or effectively eliminates the level of excitation of mOrange, and the 512/26-nanometer emission filter enables analysis of the donor fluorescence emission (Sapphire) without contamination of signal from the acceptor (mOrange). The wideband 600/80-nanometer acceptor filter collects a significant amount of signal from mOrange with less than 10 percent bleed-through of the Sapphire fluorescence. A dichromatic mirror having a cut-on wavelength of 500 nanometers (not illustrated) should be utilized with this filter combination.
Red Fluorescent Proteins
The quest for a well-behaved red-emitting fluorescent protein has long been the "holy grail" of live-cell imaging, primarily due to the requirement for probes in this spectral region in multicolor imaging experiments as well as the fact that longer excitation wavelengths generate less phototoxicity and can probe deeper into biological tissue. As an added convenience, most of the proteins in the red region of the visible spectrum can be imaged with the common TRITC and Texas Red widefield fluorescence filter sets, as well as cheap helium-neon (543, 561, 594, and 633 nanometers) lasers in confocal microscopy. After five years of unsuccessful mutagenesis efforts in the Aequorea GFP-derived proteins, the first real breakthrough occurred with the discovery of potentially fluorescent chromoproteins in non-bioluminescent Anthozoa coral species. To date, a wide spectrum of potentially useful red fluorescent proteins has been reported (spanning the emission wavelength range of 580 to 630 nanometers), many of which still suffer from some degree of the obligatory quaternary structure bestowed by their species of origin. Unlike the jellyfish proteins, most of the native and genetically engineered variants of coral reef proteins mature very efficiently at 37 degrees Celsius, presumably due to differing water temperatures of their respective native habitats.
The first Anthozoa-derived fluorescent protein to be extensively investigated was derived from the sea anemone Discosoma striata and originally referred to as drFP583, but is now commonly known as DsRed. Once the protein has fully matured, the fluorescence emission spectrum of DsRed features a peak at 583 nanometers whereas the excitation spectrum has a major peak at 558 nanometers and a minor peak around 500 nanometers. Several problems are associated with DsRed in practice. Maturation of DsRed fluorescence occurs slowly and proceeds through an intermediate chromophore stage where a majority of the fluorescence emission is seen in the green region. Termed the green state, this artifact has proven challenging for multiple labeling experiments in combination with other green fluorescent proteins because of the spectral overlap. In addition, DsRed is an obligate tetramer, an undesirable characteristic that interferes in fusion protein constructs, often leading to poor localization, and increases the tendency to form large protein aggregates in living cells. Although these side effects are not important when the probe is utilized simply as a reporter for gene expression, the utility of DsRed as an epitope tag is severely compromised. In contrast to the large Aequorea family of proteins that have been employed to successfully tag hundreds of fusion proteins, DsRed fusion proteins have proven far less successful and often exhibit toxic effects.
Several of the major problems with DsRed fluorescent protein have been overcome through site-directed and random mutagenesis efforts. The second-generation version of DsRed, known as DsRed2, contains a series of silent nucleotide substitutions corresponding to human codon preferences and several internal sequence mutations that increase the maturation rate. In addition, the elimination of basic amino acid residues (changed to acidic or neutral moieties) at the peptide amino terminus in DsRed2 reduces the tendency of the protein to form aggregates in fusion constructs. The DsRed2 protein still forms a tetramer in solution, but it is more compatible with green fluorescent proteins in multiple labeling experiments due to the increased maturation rate. Further reductions in maturation time have been realized with the third generation of DsRed mutants, which also display an increased brightness level in terms of peak cellular fluorescence. Red fluorescence emission from DsRed-Express (a commercial vector from Clontech) can be observed within an hour after expression, as compared to approximately six hours for DsRed2 and 11 hours for DsRed. The presence of a green state in DsRed-Express is not apparent, rendering this fluorescent protein the best choice of tetrameric DsRed variants for multiple labeling experiments. Because these probes remain obligate tetramers, however, they have largely been replaced with dimeric and monomeric versions and are now only of historical interest. A commercially available monomeric version of DsRed (Clontech) has recently been introduced, but the intrinsic brightness and photostability of this probe are reported to be poor.
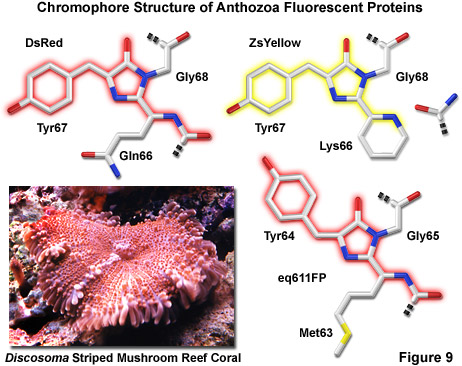
Illustrated in Figure 9 are three of the most common chromophore structural motifs exhibited by Anthozoa reef coral fluorescent proteins, including DsRed, ZsYellow and the far-red eq611FP (discussed below). The DsRed chromophore is similar in structure to that found in native green fluorescent protein, but is modified through the oxidation of a second backbone bond to extend the conjugated pi-electron system to include the phenylalanine amino acid residue at position 65 (preceding the chromophore) in the peptide chain. In addition, an unusual cis peptide bond configuration is adopted between Phe65 and Gln66, which positions the alpha-carbon atom of the glutamine residue in the same plane as the imidazolinone ring system, coinciding with the rest of the chromophore (note that GFP features a normal trans configuration at this location). The unique cis orientation of the DsRed fluorophore, which has been confirmed through crystallographic diffraction analysis, is stabilized by reduced steric hindrance, enhanced electron delocalization, and increased hydrogen bonding with neighboring amino acid residues. The presence of such an unusual bonding configuration in DsRed (which is not present in GFP) suggests that isomerization around this bond may be a key step in chromophore maturation.
Green fluorescence emission has been reported to transiently occur during formation of the ZsYellow chromophore (Figure 9) when oxidation of the tyrosine alpha-beta carbon bond by molecular oxygen extends conjugation of the imidazoline ring system to include the tyrosine phenyl ring and its para-oxygen substituent (similar to GFP). However, spectroscopic investigations have yielded evidence that the green species is formed by a dead-end route to an incomplete fluorophore product, rather than a true intermediate state. Proceeding with the maturation sequence to form the yellow fluorophore, a short-lived acylimine moiety is attacked by the terminal amino group of Lys66 to form a novel partially unsaturated six-membered piperidine ring, simultaneously cleaving the polypeptide backbone between the 65 and 66 positions. Structural analysis by x-ray diffraction indicates that the novel heterocyclic ring system lies in a plane that is approximately parallel with the rest of the ZsYellow chromophore (as illustrated in Figure 9), a finding consistent with the extended conjugation mechanism of increasing emission wavelengths. In addition, cleavage of the peptide backbone between residues Phe65 and Lys66 results in the formation of a terminal carboxamide group at residue 65, which should be available for participation in hydrogen bonding to stabilize the fluorophore.
A large number of additional red fluorescent proteins showing a considerable amount of promise have been isolated from the reef coral organisms. One of the first to be adapted for mammalian cell applications is HcRed1, which was isolated from the anemone Heteractis crispa and is now commercially available (Clontech and Evrogen). HcRed1 was originally derived from a non-fluorescent chromoprotein that absorbs orange light through site-directed and random mutagenesis. A total of six amino acid substitutions were necessary to create a red fluorescent species that matured rapidly and efficiently at 37 degrees Celsius (absorption and emission at 588 and 618 nanometers, respectively). However, similar to many other reef coral proteins, the resulting red fluorescent HcRed displays a tendency to form obligate tetramers when expressed in bacteria. Additional mutagenesis efforts resulted in a brighter dimeric variant, but a monomeric version of the protein has not yet been discovered. In order to generate a species of the protein that is useful in creating fusion products for localization studies, a tandem dimer expression vector of HcRed (two head-to-tail identical copies of the protein) has been constructed. When fused to a gene product that itself forms biopolymers (such as actin or tubulin), the HcRed tandem dimer forms an intramolecular self-association complex (mimicking a monomeric tag) that apparently does not interfere with the biological activity of the resulting chimera. However, because the overall brightness and photostability of this twinned protein combination has not yet been improved, it remains a secondary choice for routine applications in live-cell microscopy.
A far-red fluorescent protein, termed eqFP611 (Figure 9), was isolated from the sea anemone Entacmaea quadricolor and displays one of the largest Stokes shifts and red-shifted fluorescence emission wavelength profiles (excitation and emission maxima at 559 and 611 nanometers, respectively) of any naturally occurring Anthozoan fluorescent protein. The quantum yield and extinction coefficient of eqFP611 combine to yield a probe approximately as bright as EGFP. During the in vivo fluorophore maturation process, which occurs in about 12 hours, the protein passes through a green intermediate state. After maturation, however, only a small fraction of this green species (less than 1 percent) can be detected. In contrast to other Anthozoan fluorescent proteins, eqFP611 has a reduced tendency to oligomerize at lower concentrations as evidenced through electrophoresis and single-molecule experiments, although at high concentrations the protein does form tetramers. Site-directed mutagenesis efforts have yielded functional dimeric variants of eqFP611, and continued efforts may finally lead to a monomeric far-red fluorescent protein from this species.
Crystallographic studies indicate that eqFP611 forms a tetramer exhibiting 222 symmetry, which is similar to that observed for the closely related DsRed fluorescent protein and also the non-fluorescent chromoprotein Rtms5. However, within the beta-can fold, the eqFP611 fluorophore adopts a unique coplanar configuration in which the Try64 phenoxy moiety is positioned trans rather than cis (as in DsRed; see Figure 9) to the imidazoline ring system. In addition, the tri-cyclic ring system and its substituents, which probably have only limited mobility inside the protein, participate in numerous hydrogen bonds (at least nine) and a variety of non-polar van der Waals interactions with closely neighboring water molecules and amino acid residues. The extended hydrophobic methionine side chain fills a deep pocket that is also present in DsRed and Rtms5. Ultimately, the combined interactions between the fluorophore and neighboring aliphatic and aromatic amino acid side chains (and water molecules) may be helpful in elucidating the mechanism behind the unusual fluorescent properties of the eqFP611 protein.
Two additional reef coral red fluorescent proteins, AsRed2 and JRed, are commercially available (Clontech and Evrogen), but these probes form tetrameric and dimeric complexes, respectively, and are less useful than the monomeric proteins described below. AsRed2 was originally isolated as a chromoprotein from Anemonia sulcata and modified through mutagenesis to yield a protein having an absorption maximum at 576 nanometers and an emission peak at 595 nanometers with a very modest quantum yield (0.05). Although the protein has been optimized with human codons for expression in mammalian cell lines, it exhibits only about 10 percent the brightness level of EGFP and the photostability has not been reported. The dimeric protein, JRed, was derived through extensive mutagenesis of a jellyfish chromoprotein to produce a novel red fluorescent marker with peak absorption and emission wavelengths of 584 and 610 nanometers, respectively. The probe has been demonstrated to produce useful fusion tags, but is unsuitable for expression in prokaryotes due to folding problems. JRed is about 25 percent as bright as EGFP and exhibits limited photostability when illuminated in the 560 to 580 nanometers region, but can be successfully employed for long-term imaging experiments when excited with a 543-nanometer laser.
The construction of true monomeric DsRed variants, as well as monomers from proteins in other Anthozoa species, has proven to be a difficult task. A total of 33 amino acid alterations to the DsRed sequence were required for the creation of the first-generation monomeric red fluorescent protein. However, this derivative exhibits significantly reduced fluorescence emission compared to the native protein and photobleaches very quickly, rendering it much less useful than analogous monomeric green and yellow fluorescent proteins. Extensive mutagenesis research efforts, including novel techniques such as iterative somatic hypermutation, have successfully been applied in the search for yellow, orange, red, and far-red fluorescent protein variants that further reduce the tendency of these potentially efficacious biological probes to self-associate while simultaneously pushing emission maxima towards longer wavelengths. The result has been improved monomeric fluorescent proteins that feature increased extinction coefficients, quantum yields, and photostability, although no single variant has yet been optimized by all criteria. In addition, the expression problems with obligate tetrameric red fluorescent proteins are being overcome by the efforts to generate true monomeric variants, which have yielded derivatives that are more compatible with biological function.
Perhaps the most spectacular development on this front has been the introduction of a new harvest of fluorescent proteins derived from monomeric red fluorescent protein (mRFP1) through directed mutagenesis targeting the Q66 and Y67 chromophore residues, which have been demonstrated to play key roles in determining the spectral characteristics in Aequorea proteins. The resulting cadre of monomeric fluorescent proteins exhibit maxima at wavelengths ranging from 560 to 610 nanometers and have been named in honor of common fruits that bear colors similar to their respective fluorescence emission spectral profiles. Among the potentially efficacious members in the "fruit" series of fluorescent proteins are mStrawberry, mCherry, and tdTomato (a tandem dimer), all of which have fluorescence emission profiles in the orange and red regions of the spectrum. Of these probes, tdTomato, which has an orange-red emission maximum at 581 nanometers, is the brightest and most photostable under widefield illumination. However, the dimer is twice the molecular weight of the monomers. The red proteins, mCherry and mStrawberry (emission peaks at 610 and 596 nanometers, respectively), have brightness levels of approximately 50 percent and 75 percent of EGFP, but mCherry is far more photostable than mStrawberry and is the best probe choice to replace mRFP1 for long-term imaging experiments. These new proteins essentially fill the gap between the most red-shifted jellyfish fluorescent proteins (such as Venus) and the multitude of oligomeric coral reef red fluorescent proteins that have been reported and are commercially available. Although several of these new fluorescent monomeric proteins lack the brightness and photostability necessary for many imaging experiments, their existence is encouraging as it suggests the eventuality of bright, stable, monomeric fluorescent proteins across the entire visible spectrum.
Far-Red Fluorescent Proteins
Further extension of the fruit protein spectral class through iterative somatic hypermutation has yielded two new fluorescent proteins with emission wavelength maxima of 625 and 649 nanometers, representing the first true far-red (ranging from 630 to 700 nanometers) genetically engineered probes. The most potentially useful probe in this pair was named mPlum, which has a rather limited brightness value (10 percent of EGFP), but excellent photostability. This monomeric probe should be useful in combination with fluorescent proteins emitting in the cyan, green, yellow, and orange regions for multicolor imaging experiments (see Figure 11) and as a biosensor FRET partner with green and yellow proteins, such as mEmerald (Förster distance, 4.4) and mCitrine (Förster distance, 5.0). Another far-red fluorescent protein, termed AQ143, has been derived from mutagenesis efforts on a chromoprotein isolated from the anemone Actinia equine. The excitation and emission maxima of AQ143 are 595 and 655 nanometers, respectively, and the brightness is comparable to mPlum. On the downside, the photostability of this protein has not been reported and it forms an obligate tetramer.
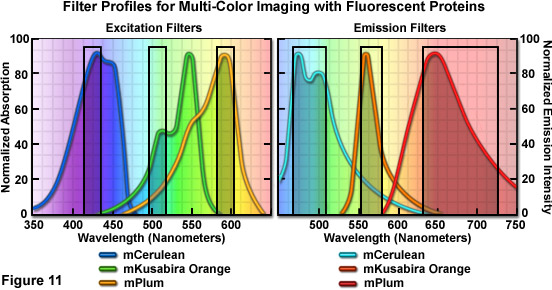
Optimized fluorescence filter combinations for multicolor imaging of three fluorescent proteins spanning the cyan through far-red wavelength regions are presented in Figure 11. The excitation filters are optimized for Cerulean, mKO, and mPlum fluorescent proteins having center wavelengths of 425, 508, and 585 nanometers, respectively. The bandwidth of all excitation filters is 20 nanometers. The emission filters are optimized for the same probes having center wavelengths of 480, 564, and 675 nanometers with bandwidths of 40, 28, and 130 nanometers, respectively. The mKO excitation filter is designed to work off-peak to minimize simultaneous excitation of mPlum. Bleed-through of Cerulean fluorescence emission into the mKO filter is reduced by the fact that mKO is four times brighter at equimolar concentrations and requires much shorter exposure intervals for image capture.
Conclusions
The primary thrust of fluorescent protein development is centered on fine-tuning the current palette of blue to yellow fluorescent proteins derived from the Aequorea victoria jellyfish, while simultaneously developing monomeric fluorescent proteins emitting in the orange to far-red regions of the visible light spectrum. Progress toward these goals has been substantial, and it is not inconceivable that near-infrared emitting fluorescent proteins loom on the horizon. The latest efforts in jellyfish variants have resulted in new and improved monomeric probes for the cyan, green, and yellow region, while the search for a bright, monomeric, and fast-maturing red fluorescent protein has yielded a host of promising candidates that extend into the longer wavelength regions. Continued protein engineering of the existing fluorescent proteins coupled with new technologies, such as the application of unnatural amino acids and circular permutation, should further expand the color palette.
The complex interplay of biological roles for an ever-increasing number of fluorescent proteins derived from a wide variety of marine species is only beginning to be understood. Light-induced changes to the autocatalytic chromophores, including photoactivation and photoconversion, may serve as a highly evolved photo-protection mechanism to assist these organisms in the useful dissipation of high-energy sunlight, especially the damaging shorter wavelengths, via the absorption and subsequent fluorescence re-emission of longer and safer wavelengths. In many cases, these remarkable fluorescent proteins display very high photostability and dynamic photo-induced transformation properties, including spectral fine-tuning of donor-acceptor pairs (and even cascades) for resonance energy transfer. The large number of spectral variants already discovered, featuring emission profiles covering the entire visible spectrum, suggests that the diverse optical and biochemical properties of these proteins will generate a host of new candidates as probes for biological investigations and ensure the continued development of unique genetically engineered fluorescent proteins.
