Fiber FISH (Fluorescence in situ Hybridization)
The term Fiber FISH refers to the common practice of fluorescence in situ hybridization (FISH) conducted on preparations of extended chromatin fibers. In mapping DNA fragments of interest by conducting FISH investigations on chromosomes, signals within a distance of several million base pairs are indistinguishable from each other because of the multifold structure of DNA strands in the metaphase chromosomes. The resolution of signals improves if the chromosomes are used before they progress to full condensation. What can we do when we want to map more adjacent DNA clones? The characterization of entire genome DNA sequences will resolve the problem of creating a map in scale of one base pair, but is extremely time-consuming.
The human genome is calculated to contain 30,000-100,000 genes, that is, 1,200-4,000 genes per chromosome on average. The genes of 10-15 thousand base pairs of average size line up on DNA strands at intervals of 40-45 thousand base pairs. New procedures are required in order to produce a detailed map of DNA fragments containing these genes.
The following discussion addresses two mechanisms to overcome the resolution limits encountered when using FISH on metaphase chromosomes. Mapping segments at under one million base pairs (MBPs) resolution is available by using stretched chromatin (DNA) fibers.
Mapping Under One Million Base Pairs (MBPs) Resolution
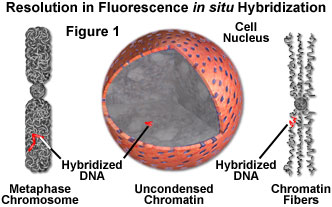
- FISH on Interphase Nuclei - A high-resolution map can be created by using the interphase nuclei as chromatin fibers extending into the nuclei because they stretch much more when compared to those in the metaphase chromosomes (see Figure 1). This procedure is can be used to map DNA clones at a resolution of 50-500 kilobase pairs, however its accuracy is somewhat reduced because the degree of extension of chromatin fibers varies.
- FISH on Extended Chromatin Fiber Preparation - A higher resolution method for detection of signals is obtained by artificially extending chromatin fibers on a slide glass before DNA hybridization (Figure 1).
Fiber FISH Procedure
- Cells are spread and air-dried on a slide glass.
- The cells on a slide glass are lysed by detergent (Triton X-100) in a closed container.
- Chromatin fibers attach to the slide glass when it is slowly removed from the container.
- The preparation is fixed with ethanol.
- High resolution FISH at a distance of 5-700 kilobase pairs becomes possible.
- The probes are hybridized following standard procedures.
Fiber FISH is currently used to order a series of DNA fragments cloned from chromosomes and to estimate the distance between these clones. However, DNA strands must still be examined at the submicroscopic level for more detail. The difficulty of submicroscopic measurement makes tracking and distance measurements unclear. Current research is exploring potential methods of simultaneous detection of signals and DNA strands. Scanning probe microscopy is a promising candidate.
Internet Resources
- Fiber FISH Images - Presented by SeeDNA Biotech Inc., these fiber FISH images demonstrate the enhanced resolution obtainable with the technology.
